what happens to the chloride gradient in CF
| CFTR | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
  | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||
| Aliases | CFTR, ABC35, ABCC7, CF, CFTR/MRP, MRP7, TNR-dJ760C5.i, cystic fibrosis transmembrane conductance regulator, CF transmembrane conductance regulator | ||||||||||||||||||||||||
| External IDs | OMIM: 602421 MGI: 88388 HomoloGene: 55465 GeneCards: CFTR | ||||||||||||||||||||||||
| EC number | 5.half-dozen.1.half dozen | ||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| |||||||||||||||||||||||||
| Orthologs | |||||||||||||||||||||||||
| Species | Human | Mouse | |||||||||||||||||||||||
| Entrez |
|
| |||||||||||||||||||||||
| Ensembl |
|
| |||||||||||||||||||||||
| UniProt |
|
| |||||||||||||||||||||||
| RefSeq (mRNA) |
|
| |||||||||||||||||||||||
| RefSeq (poly peptide) |
|
| |||||||||||||||||||||||
| Location (UCSC) | Chr 7: 117.29 – 117.72 Mb | Chr 6: 18.17 – 18.32 Mb | |||||||||||||||||||||||
| PubMed search | [3] | [iv] | |||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||
| |||||||||||||||||||||||||
Cystic fibrosis transmembrane conductance regulator (CFTR) is a membrane protein and chloride channel in vertebrates that is encoded by the CFTR gene.[5] [6]
The CFTR gene codes for an ABC transporter-class ion aqueduct protein that conducts chloride[seven] ions across epithelial cell membranes. Mutations of the CFTR gene affecting chloride ion channel function pb to dysregulation of epithelial fluid send in the lung, pancreas and other organs, resulting in cystic fibrosis. Complications include thickened mucus in the lungs with frequent respiratory infections, and pancreatic insufficiency giving rise to malnutrition and diabetes. These conditions lead to chronic inability and reduced life expectancy. In male patients, the progressive obstruction and destruction of the developing vas deferens (spermatic cord) and epididymis announced to event from abnormal intraluminal secretions,[8] causing congenital absence of the vas deferens and male infertility.
Gene [edit]

The location of the CFTR gene on chromosome 7
The factor that encodes the human CFTR protein is found on chromosome vii, on the long arm at position q31.2.[6] from base pair 116,907,253 to base pair 117,095,955. CFTR orthologs[9] occur in the jawed vertebrates.[10]
The CFTR cistron has been used in animals as a nuclear DNA phylogenetic marker.[nine] Big genomic sequences of this gene accept been used to explore the phylogeny of the major groups of mammals,[11] and confirmed the grouping of placental orders into four major clades: Xenarthra, Afrotheria, Laurasiatheria, and Euarchonta plus Glires.
Mutations [edit]
Near 1000 cystic fibrosis-causing mutations have been described.[12] The nigh common mutation, DeltaF508 (ΔF508) results from a deletion (Δ) of three nucleotides which results in a loss of the amino acrid phenylalanine (F) at the 508th position on the protein.[xiii] As a consequence, the poly peptide does not fold normally and is more quickly degraded. The vast majority of mutations are infrequent. The distribution and frequency of mutations varies among different populations which has implications for genetic screening and counseling.
Drug discovery for therapeutics to accost CF in all patients is complicated due to the large number of illness-causing mutations. Ideally, a library of cell lines and prison cell-based assays corresponding to all mutants is required to screen for broadly-active drug candidates. Prison cell applied science methods including fluorogenic oligonucleotide signaling probes may be used to detect and isolate clonal cell lines for each mutant.[xiv]
Mutations consist of replacements, duplications, deletions or shortenings in the CFTR factor. This may issue in proteins that may not function, piece of work less effectively, are more than speedily degraded, or are nowadays in inadequate numbers.[15]
It has been hypothesized that mutations in the CFTR factor may confer a selective advantage to heterozygous individuals. Cells expressing a mutant form of the CFTR protein are resistant to invasion by the Salmonella typhi bacterium, the agent of typhoid fever, and mice carrying a single copy of mutant CFTR are resistant to diarrhea acquired by cholera toxin.[16]
The virtually common mutations that cause cystic fibrosis and pancreatic insufficiency in humans are:[17]
| Variant cDNA name (ordered 5' to iii') | Variant poly peptide name | Variant legacy name | rsID | # alleles in CFTR2 | Allele frequency in CFTR2 | % pancreatic insufficient | Variant final determination (July 2020) |
| c.1521_1523delCTT | p.Phe508del | F508del | rs113993960 | 99061 | 0.69744 | 98% | CF-causing |
| c.1624G>T | p.Gly542X | G542X | rs113993959 | 3610 | 0.02542 | 98% | CF-causing |
| c.1652G>A | p.Gly551Asp | G551D | rs75527207 | 2986 | 0.02102 | 96% | CF-causing |
| c.3909C>Chiliad | p.Asn1303Lys | N1303K | rs80034486 | 2246 | 0.01581 | 98% | CF-causing |
| c.350G>A | p.Arg117His | R117H | rs78655421 | 1854 | 0.01305 | 23% | Varying clinical consequence |
| c.3846G>A | p.Trp1282X | W1282X | rs77010898 | 1726 | 0.01215 | 99% | CF-causing |
| c.489+1G>T | No protein name | 621+1G->T | rs78756941 | 1323 | 0.00931 | 99% | CF-causing |
| c.1657C>T | p.Arg553X | R553X | rs74597325 | 1323 | 0.00931 | 97% | CF-causing |
| c.1585-1G>A | No poly peptide name | 1717-1G->A | rs76713772 | 1216 | 0.00856 | 97% | CF-causing |
| c.3718-2477C>T | No poly peptide proper name | 3849+10kbC->T | rs75039782 | 1158 | 0.00815 | 33% | CF-causing |
| c.2657+5G>A | No protein name | 2789+5G->A | rs80224560 | 1027 | 0.00723 | 43% | CF-causing |
| c.1519_1521delATC | p. Ile507del | I507del | rs121908745 | 651 | 0.00458 | 98% | CF-causing |
| c.3484C>T | p.Arg1162X | R1162X | rs74767530 | 651 | 0.00458 | 97% | CF-causing |
| c.254G>A | p.Gly85Glu | G85E | rs75961395 | 616 | 0.00434 | 85% | CF-causing |
| c.3454G>C | p.Asp1152His | D1152H | rs75541969 | 571 | 0.00402 | 24% | Varying clinical consequence |
| c.2051_2052delAAinsG | p. Lys684SerfsX38 | 2183AA->G | rs121908799 | 542 | 0.00382 | 96% | CF-causing |
| c.3528delC | p. Lys1177SerfsX15 | 3659delC | rs121908747 | 539 | 0.00379 | 99% | CF-causing |
| c.1040G>C | p.Arg347Pro | R347P | rs77932196 | 533 | 0.00375 | 68% | CF-causing |
| c.1210−12T[5] | No protein name | 5T | rs1805177 | 516 | 0.00363 | 28% | Varying clinical consequence |
| c.2988+1G>A | No protein name | 3120+1G->A | rs75096551 | 501 | 0.00353 | 98% | CF-causing |
| c.1364C>A | p.Ala455Glu | A455E | rs74551128 | 500 | 0.00352 | 34% | CF-causing |
| c.3140-26A>G | No protein name | 3272-26A->Chiliad | rs76151804 | 470 | 0.00331 | 29% | CF-causing |
| c.1000C>T | p.Arg334Trp | R334W | rs121909011 | 429 | 0.00302 | 40% | CF-causing |
| c.1766+1G>A | No protein proper name | 1898+1G->A | rs121908748 | 421 | 0.00296 | 99% | CF-causing |
| c.54-5940_273+10250del21kb | p.Ser18ArgfsX16 | CFTRdele2,3 | not found | 417 | 0.00294 | 100% | CF-causing |
| c.1679G>C | p.Arg560Thr | R560T | rs80055610 | 343 | 0.00241 | 98% | CF-causing |
| c.617T>G | p. Leu206Trp | L206W | rs121908752 | 333 | 0.00234 | 20% | CF-causing |
| c.2052dupA | p.Gln685ThrfsX4 | 2184insA | rs121908786 | 329 | 0.00232 | 85% | CF-causing |
| c.262_263delTT | p. Leu88IlefsX22 | 394delTT | rs121908769 | 307 | 0.00216 | 97% | CF-causing |
| c.178G>T | p.Glu60X | E60X | rs77284892 | 296 | 0.00208 | 99% | CF-causing |
| c.1477C>T | p.Gln493X | Q493X | rs77101217 | 292 | 0.00206 | 98% | CF-causing |
| c.579+1G>T | No protein proper noun | 711+1G->T | rs77188391 | 274 | 0.00193 | 98% | CF-causing |
| c.2052delA | p. Lys684AsnfsX38 | 2184delA | rs121908746 | 255 | 0.00180 | 98% | CF-causing |
| c.200C>T | p.Pro67Leu | P67L | rs368505753 | 239 | 0.00168 | 34% | CF-causing |
| c.3302T>A | p.Met1101Lys | M1101K | rs36210737 | 238 | 0.00168 | 69% | CF-causing |
| c.1408A>G | p.Met470Val | M470V | rs213950 | 235 | 0.00165 | 46% | Non CF-causing |
| c.3276C>A or c.3276C>K | p.Tyr1092X | Y1092X | rs121908761 | 225 | 0.00158 | 98% | CF-causing |
| c.3196C>T | p.Arg1066Cys | R1066C | rs78194216 | 220 | 0.00155 | 98% | CF-causing |
| c.1021_1022dupTC | p.Phe342HisfsX28 | 1154insTC | rs387906360 | 214 | 0.00151 | 99% | CF-causing |
| c.3773dupT | p. Leu1258PhefsX7 | 3905insT | rs121908789 | 210 | 0.00148 | 97% | CF-causing |
| c.1646G>A | p.Ser549Asn | S549N | rs121908755 | 203 | 0.00143 | 84% | CF-causing |
| c.1040G>A | p.Arg347His | R347H | rs77932196 | 199 | 0.00140 | 24% | CF-causing |
| c.948delT | p.Phe316LeufsX12 | 1078delT | rs121908744 | 184 | 0.00130 | 99% | CF-causing |
| c.1210-33_1210-6GT[12]T[4] | No protein proper noun | 5T;TG12 | not plant | 182 | 0.00128 | fourteen% | Varying clinical consequence |
| c.3472C>T | p.Arg1158X | R1158X | rs79850223 | 179 | 0.00126 | 99% | CF-causing |
| c.2834C>T | p.Ser945Leu | S945L | rs397508442 | 167 | 0.00118 | 40% | CF-causing |
| c.1558G>T | p. Val520Phe | V520F | rs77646904 | 156 | 0.00110 | 98% | CF-causing |
| c.443T>C | p. Ile148Thr | I148T | rs35516286 | 148 | 0.00104 | 88% | Not CF-causing |
| c.349C>T | p.Arg117Cys | R117C | rs77834169 | 146 | 0.00103 | 24% | CF-causing |
DeltaF508 [edit]
DeltaF508 (ΔF508), full proper noun CFTRΔF508 or F508del-CFTR (rs113993960), is a specific mutation within the CFTR factor involving a deletion of 3 nucleotides spanning positions 507 and 508 of the CFTR gene on chromosome 7, which ultimately results in the loss of a single codon for the amino acid phenylalanine (F). A person with the CFTRΔF508 mutation will produce an abnormal CFTR poly peptide that lacks this phenylalanine residue and which cannot fold properly. This poly peptide does non escape the endoplasmic reticulum for further processing. Having two copies of this mutation (one inherited from each parent) is by far the well-nigh common cause of cystic fibrosis (CF), responsible for near two-thirds of mutations worldwide.[18]
Effects [edit]
The CFTR protein is largely expressed in cells of the pancreas, intestinal and respiratory epithelia, and all exocrine glands. When properly folded, it is shuttled to the prison cell membrane, where it becomes a transmembrane poly peptide responsible for opening channels which release chloride ions out of cells; it also simultaneously inhibits the uptake of sodium ions past another aqueduct poly peptide. Both of these functions help to maintain an ion slope that causes osmosis to draw h2o out of the cells.[19] The ΔF508 mutation leads to the misfolding of CFTR and its eventual degradation in the ER. In organisms with 2 complements of the mutation, the protein is entirely absent from the prison cell membrane, and these critical ion ship functions are not performed.[20]
Having a homozygous pair of genes with the ΔF508 mutation prevents the CFTR protein from assuming its normal position in the jail cell membrane. This causes increased h2o retention in cells, respective dehydration of the extracellular space, and an associated cascade of effects on various parts of the body. These effects include: thicker mucous membranes in the epithelia of afflicted organs; obstruction of narrow respiratory airways as a issue of thicker mucous and inhibition of the free movement of mucocilia; congenital absenteeism of the vas deferens due to increased fungus thickness during fetal evolution; pancreatic insufficiency due to blockage of the pancreatic duct with mucus; and increased risk of respiratory infection due to build-up of thick, food-rich mucus where bacteria thrive. These are the symptoms of cystic fibrosis, a genetic disorder; however, ΔF508 is not the only mutation that causes this disorder.
Beingness a heterozygous carrier (having a single copy of ΔF508) results in decreased water loss during diarrhea because malfunctioning or absent CFTR proteins cannot maintain stable ion gradients across cell membranes. Typically in that location is a build-up of both Cl− and Na+ ions within affected cells, creating a hypotonic solution outside the cells and causing water to diffuse into the cells by osmosis. Several studies indicate that heterozygous carriers are at increased risk for various symptoms. For case, it has been shown that heterozygosity for cystic fibrosis is associated with increased airway reactivity, and heterozygotes may be at risk for poor pulmonary function. Heterozygotes with wheeze have been shown to be at college gamble for poor pulmonary role or evolution and progression of chronic obstructive lung illness. Ane gene for cystic fibrosis is sufficient to produce balmy lung abnormalities even in the absence of infection.[21]
Mechanism [edit]
The CFTR gene is located on the long arm of chromosome seven, at position q31.ii, and ultimately codes for a sequence of 1,480 amino acids. Normally, the three DNA base pairs A-T-C (paired with T-A-G on the opposite strand) at the gene'southward 507th position form the template for the mRNA codon A-U-C for isoleucine, while the three DNA base pairs T-T-T (paired with A-A-A) at the next 508th position course the template for the codon U-U-U for phenylalanine.[22] The ΔF508 mutation is a deletion of the C-G pair from position 507 along with the get-go ii T-A pairs from position 508, leaving the Dna sequence A-T-T (paired with T-A-A) at position 507, which is transcribed into the mRNA codon A-U-U. Since A-U-U too codes for isoleucine, position 507'due south amino acrid does non alter, and the mutation's net effect is equivalent to a deletion ("Δ") of the sequence resulting in the codon for phenylalanine at position 508.[23]
Prevalence [edit]
ΔF508 is present on at least one copy of chromosome 7 in approximately one in 30 Caucasians. Presence of the mutation on both copies causes the autosomal recessive disease cystic fibrosis. Scientists accept estimated that the original mutation occurred over 52,000 years ago in Northern Europe. The young allele historic period may be a consequence of past selection. One hypothesis as to why the otherwise detrimental mutation has been maintained past natural selection is that a single copy may present a positive upshot past reducing h2o loss during cholera, though the introduction of pathogenic Vibrio cholerae into Europe did not occur until the late 18th century.[24] Another theory posits that CF carriers (heterozygotes for ΔF508) are more resistant to typhoid fever, since CFTR has been shown to act as a receptor for Salmonella typhi bacteria to enter abdominal epithelial cells.[25]
Cystic fibrosis ΔF508 heterozygotes may be overrepresented among individuals with asthma and may have poorer lung office than non-carriers.[26] [27] Carriers of a single CF mutation have a higher prevalence of chronic rhinosinusitis than the general population.[28] Approximately 50% of cystic fibrosis cases in Europe are due to homozygous ΔF508 mutations (this varies widely by region),[29] while the allele frequency of ΔF508 is well-nigh 70%.[xxx] The remaining cases are caused by over 1,500 other mutations, including R117H, 1717-1G>A, and 2789+56G>A. These mutations, when combined with each other or even a unmarried copy of ΔF508, may cause CF symptoms. The genotype is non strongly correlated with severity of the CF, though specific symptoms have been linked to sure mutations.
Structure [edit]

The Overall Structure of Man CFTR in the Dephosphorylated, ATP-Free Conformation. Domains are labeled. Made from PDB 5UAK [1]
The CFTR cistron is approximately 189 kb in length, with 27 exons and 26 introns.[31] CFTR is a glycoprotein with 1480 amino acids. The protein consists of 7 domains. There are 2 transmembrane domains, each with vi spans of alpha helices. These are each continued to a nucleotide binding domain (NBD) in the cytoplasm. The get-go NBD is connected to the second transmembrane domain by a regulatory "R" domain that is a unique feature of CFTR, non nowadays in other ABC transporters. The ion channel only opens when its R-domain has been phosphorylated by PKA and ATP is bound at the NBDs.[32] The amino-terminus is part of the lasso motif which anchors into the cell membrane.[33] The carboxyl terminal of the poly peptide is anchored to the cytoskeleton by a PDZ-interacting domain.[34] The construction shown (PDB# 1XMI) shows a homopentameric assembly of mutated NBD1, the showtime nucleotide binding domain (NBD1) of the transporter.
Location and function [edit]
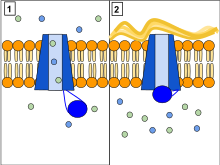
The CFTR protein is a channel protein that controls the flow of H2O and Cl− ions in and out of cells inside the lungs. When the CFTR poly peptide is working correctly, as shown in Panel 1, ions freely flow in and out of the cells. However, when the CFTR poly peptide is malfunctioning every bit in Panel two, these ions cannot catamenia out of the cell due to blocked CFTR channels. This occurs in cystic fibrosis, characterized by the buildup of thick mucus in the lungs.
CFTR functions as a phosphorylation and ATP-gated anion channel, increasing the conductance for sure anions (due east.g. Cl−) to menstruation downward their electrochemical gradient. ATP-driven conformational changes in CFTR open and close a gate to allow transmembrane flow of anions downwards their electrochemical slope.[5] This in contrast to other ABC proteins, in which ATP-driven conformational changes fuel uphill substrate ship across cellular membranes. Essentially, CFTR is an ion channel that evolved as a 'broken' ABC transporter that leaks when in open conformation.
CFTRs have ii transmembrane domains, each linked to a nucleotide-bounden domain. CFTR also contains another domain called the regulatory domain. Other members of the ABC transporter superfamily are involved in the uptake of nutrients in prokaryotes, or in the export of a variety of substrates in eukaryotes. ABC transporters accept evolved to transduce the gratis energy of ATP hydrolysis to the uphill movement of substrates beyond the cell membrane. They have ii main conformations, i where the cargo binding site is facing the cytosol or inwards facing (ATP free), and one where it is outward facing (ATP bound). ATP binds to each nucleotide binding domain, which results in the subsequent NBD dimerization, leading to the rearrangement of the transmembrane helices. This changes the accessibility of the cargo binding site from an inward facing position to an outward facing ane. ATP binding, and the hydrolysis that follows, drives the culling exposure of the cargo binding site, ensuring a unidirectional transport of cargo confronting an electrochemical gradient. In CFTR, alternating between an inwards-facing conformation to an outward-facing i results in channel gating. In item, NBD dimerization (favoured by ATP binding) is coupled to transition to an outward-facing conformation in which an open transmembrane pathway for anions is formed. Subsequent hydrolysis (at the approved agile site, site 2, including Walker motifs of NBD2) destabilizes the NBD dimer and favours return to the inward-facing conformation, in which the anion permeation pathway is airtight off.[five]
The CFTR is found in the epithelial cells of many organs including the lung, liver, pancreas, digestive tract, and the female person[35] and male reproductive tracts.[36] [37]
In the airways of the lung, CFTR is near highly expressed by rare specialized cells called pulmonary ionocytes.[38] [39] [40] In the pare CFTR is strongly expressed in the sebaceous and eccrine sweat glands.[41] In the eccrine glands, CFTR is located on the apical membrane of the epithelial cells that make up the duct of these sweat glands.[41]
Ordinarily, the protein allows motility of chloride and thiocyanate[42] ions (with a negative accuse) out of an epithelial cell into the Airway Surface Liquid and mucus. Positively charged sodium ions follow passively, increasing the total electrolyte concentration in the mucus, resulting in the movement of water out of the cell via osmosis.
In epithelial cells with motile cilia lining the bronchus and the oviduct, CFTR is located on the apical cell membrane but not on cilia.[35] In dissimilarity, ENaC (Epithelial sodium channel) is located along the unabridged length of the cilia.[35]
In sweat glands, defective CFTR results in reduced transport of sodium chloride and sodium thiocyanate[43] in the reabsorptive duct and therefore saltier sweat. This is the footing of a clinically important sweat examination for cystic fibrosis oft used diagnostically with genetic screening.[44]
Interactions [edit]
Cystic fibrosis transmembrane conductance regulator has been shown to interact with:
- DNAJC5,[45]
- GOPC,[46] [47]
- PDZK1,[47] [48]
- PRKCE,[49]
- SLC4A8,[fifty]
- SNAP23,[51]
- SLC9A3R1,[34] [50] [52] [53] [54] [55]
- SLC9A3R2,[56] and
- STX1A,[51] [57]
It is inhibited by the anti-diarrhoea drug crofelemer.
[edit]
- Congenital bilateral absence of vas deferens: Males with congenital bilateral absenteeism of the vas deferens most frequently have a mild mutation (a alter that allows partial function of the gene) in i copy of the CFTR gene and a cystic fibrosis-causing mutation in the other copy of CFTR.
- Cystic fibrosis: More 1,800 mutations in the CFTR gene take been found[58] merely the bulk of these take not been associated with cystic fibrosis.[59] Most of these mutations either substitute i amino acid (a building block of proteins) for some other amino acid in the CFTR protein or delete a pocket-size amount of Deoxyribonucleic acid in the CFTR cistron. The virtually common mutation, chosen ΔF508, is a deletion (Δ) of one amino acrid (phenylalanine) at position 508 in the CFTR protein. This altered protein never reaches the cell membrane because it is degraded shortly subsequently it is fabricated. All affliction-causing mutations in the CFTR gene prevent the aqueduct from performance properly, leading to a blockage of the motion of table salt and h2o into and out of cells. As a result of this blockage, cells that line the passageways of the lungs, pancreas, and other organs produce abnormally thick, glutinous mucus. This mucus obstructs the airways and glands, causing the characteristic signs and symptoms of cystic fibrosis. In addition, only thin fungus can be removed past cilia; thick mucus cannot, and so it traps bacteria that give rise to chronic infections.
- Cholera: ADP-ribosylation caused by cholera toxin results in increased production of cyclic AMP which in plough opens the CFTR channel which leads to oversecretion of Cl−. Na+ and H2O follow Cl− into the small intestine, resulting in dehydration and loss of electrolytes.[lx]
Drug target [edit]
CFTR has been a drug target in efforts to find treatments for related conditions. Ivacaftor (merchandise proper noun Kalydeco, adult equally VX-770) is a drug approved by the FDA in 2012 for people with cystic fibrosis who accept specific CFTR mutations.[61] [62] Ivacaftor was adult by Vertex Pharmaceuticals in conjunction with the Cystic Fibrosis Foundation and is the starting time drug that treats the underlying crusade rather than the symptoms of the disease.[63] Chosen "the virtually important new drug of 2012",[64] and "a wonder drug"[65] information technology is one of the most expensive drugs, costing over U.s.a.$300,000 per twelvemonth, which has led to criticism of Vertex for the high price.
References [edit]
- ^ a b c GRCh38: Ensembl release 89: ENSG00000001626 - Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000041301 - Ensembl, May 2017
- ^ "Human being PubMed Reference:". National Centre for Biotechnology Data, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b c Csanády L, Vergani P, Gadsby DC (January 2019). "Structure, Gating, and Regulation of the CFTR Anion Channel" (PDF). Physiological Reviews. 99 (1): 707–738. doi:ten.1152/physrev.00007.2018. PMID 30516439.
- ^ a b Collins F, Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer K, et al. (September 1989). "Identification of the cystic fibrosis factor: chromosome walking and jumping". Science. 245 (4922): 1059–65. Bibcode:1989Sci...245.1059R. doi:10.1126/science.2772657. PMID 2772657.
- ^ Riordan JR, Rommens JM, Kerem B, Alon North, Rozmahel R, Grzelczak Z, et al. (September 1989). "Identification of the cystic fibrosis gene: cloning and label of complementary DNA". Science. 245 (4922): 1066–73. Bibcode:1989Sci...245.1066R. doi:10.1126/science.2475911. PMID 2475911.
- ^ Marcorelles P, Gillet D, Friocourt G, Ledé F, Samaison 50, Huguen G, Ferec C (March 2012). "Cystic fibrosis transmembrane conductance regulator protein expression in the male excretory duct system during evolution". Human Pathology. 43 (3): 390–7. doi:10.1016/j.humpath.2011.04.031. PMID 21840567.
- ^ a b "OrthoMaM phylogenetic marker: CFTR coding sequence". Archived from the original on 2016-03-02. Retrieved 2010-03-12 .
- ^ Davies R, Conroy SJ, Davies WL, Potter IC, Trezise AE (19–23 June 2005). "Evolution and Regulation of the Cystic Fibrosis Cistron" (conference newspaper). Molecular Biology and Evolution (MBE05) Conference . Retrieved 28 July 2014.
- ^ Prasad AB, Allard MW, Light-green ED (September 2008). "Confirming the phylogeny of mammals by use of large comparative sequence data sets". Molecular Biology and Evolution. 25 (ix): 1795–808. doi:x.1093/molbev/msn104. PMC2515873. PMID 18453548.
- ^ "The Clinical and Functional TRanslation of CFTR (CFTR2): CFTR2 Variant List History". US CF Foundation, Johns Hopkins University, Cystic Fibrosis Centre at the Hospital for Sick Children in Toronto. Retrieved 2 August 2017. [ permanent expressionless link ]
- ^ Guimbellot J, Sharma J, Rowe SM (November 2017). "Toward inclusive therapy with CFTR modulators: Progress and challenges". Pediatric Pulmonology. 52 (S48): S4–S14. doi:10.1002/ppul.23773. PMC6208153. PMID 28881097.
- ^ Shekdar One thousand, Langer J, Venkatachalan S, Schmid L, Anobile J, Shah P, et al. (March 2021). "Cell engineering method using fluorogenic oligonucleotide signaling probes and flow cytometry". Biotechnology Letters. 43 (5): 949–958. doi:x.1007/s10529-021-03101-v. PMC7937778. PMID 33683511.
- ^ Rowe SM, Miller S, Sorscher EJ (May 2005). "Cystic fibrosis". The New England Journal of Medicine. 352 (nineteen): 1992–2001. doi:x.1056/NEJMra043184. PMID 15888700.
- ^ Kavic SM, Frehm EJ, Segal AS (1999). "Example studies in cholera: lessons in medical history and scientific discipline". The Yale Journal of Biology and Medicine. 72 (6): 393–408. PMC2579035. PMID 11138935.
- ^ "CFTR2". Retrieved 2021-07-08 .
- ^ Bobadilla JL, Macek M, Fine JP, Farrell PM (June 2002). "Cystic fibrosis: a worldwide assay of CFTR mutations--correlation with incidence information and application to screening". Human being Mutation. 19 (six): 575–606. doi:10.1002/humu.10041. PMID 12007216.
- ^ Verkman Every bit, Song Y, Thiagarajah JR (January 2003). "Role of airway surface liquid and submucosal glands in cystic fibrosis lung disease". American Journal of Physiology. Cell Physiology. 284 (1): C2-15. doi:x.1152/ajpcell.00417.2002. PMID 12475759.
- ^ "Cystic Fibrosis Research Directions". National Found of Diabetes and Digestive and Kidney Diseases (NIDDK).
- ^ Maurya N, Awasthi Due south, Dixit P (April 2012). "Association of CFTR factor mutation with bronchial asthma" (PDF). The Indian Journal of Medical Enquiry. 135 (4): 469–78. PMC3385229. PMID 22664493.
- ^ CCDS Written report for Consensus CDS: Report for CCDS5773.1 (current version) NCBI
- ^ Bartoszewski RA, Jablonsky M, Bartoszewska S, Stevenson Fifty, Dai Q, Kappes J, et al. (September 2010). "A synonymous unmarried nucleotide polymorphism in DeltaF508 CFTR alters the secondary structure of the mRNA and the expression of the mutant protein". The Journal of Biological Chemistry. 285 (37): 28741–8. doi:10.1074/jbc.M110.154575. PMC2937902. PMID 20628052.
- ^ "Re: Is at that place a connexion between cystic fibrosis and cholera?".
- ^ Pier GB, Grout M, Zaidi T, Meluleni Grand, Mueschenborn SS, Banting G, et al. (May 1998). "Salmonella typhi uses CFTR to enter intestinal epithelial cells". Nature. 393 (6680): 79–82. Bibcode:1998Natur.393...79P. doi:10.1038/30006. PMID 9590693. S2CID 5894247.
- ^ Dahl M, Nordestgaard BG, Lange P, Tybjaerg-Hansen A (May 2001). "Xv-year follow-upwardly of pulmonary office in individuals heterozygous for the cystic fibrosis phenylalanine-508 deletion". The Journal of Allergy and Clinical Immunology. 107 (5): 818–23. doi:x.1067/mai.2001.114117. PMID 11344348.
- ^ Dahl M, Tybjaerg-Hansen A, Lange P, Nordestgaard BG (June 1998). "DeltaF508 heterozygosity in cystic fibrosis and susceptibility to asthma". Lancet. 351 (9120): 1911–3. doi:10.1016/s0140-6736(97)11419-two. PMID 9654257. S2CID 22970136.
- ^ Wang X, Kim J, McWilliams R, Cutting GR (March 2005). "Increased prevalence of chronic rhinosinusitis in carriers of a cystic fibrosis mutation". Athenaeum of Otolaryngology–Head & Neck Surgery. 131 (three): 237–40. doi:x.1001/archotol.131.3.237. PMID 15781764.
- ^ ECFS Annual Report: What It Means to the U.k. Cystic Fibrosis Trust
- ^ Morral North, Bertranpetit J, Estivill Ten, Nunes V, Casals T, Giménez J, et al. (June 1994). "The origin of the major cystic fibrosis mutation (delta F508) in European populations". Nature Genetics. seven (2): 169–75. doi:10.1038/ng0694-169. PMID 7920636. S2CID 38005421.
- ^ Cystic Fibrosis Mutation Database. "Genomic Deoxyribonucleic acid sequence". Archived from the original on 2016-08-22. Retrieved 2013-04-06 .
- ^ Sheppard DN, Welsh MJ (January 1999). "Construction and part of the CFTR chloride channel". Physiological Reviews. 79 (1 Suppl): S23-45. doi:10.1152/physrev.1999.79.1.S23. PMID 9922375.
- ^ Zhang Z, Liu F, Chen J (December 2018). "Molecular structure of the ATP-jump, phosphorylated homo CFTR". Proceedings of the National Academy of Sciences of the United states of america. 115 (fifty): 12757–12762. doi:x.1073/pnas.1815287115. PMC6294961. PMID 30459277.
- ^ a b Brusk DB, Trotter KW, Reczek D, Kreda SM, Bretscher A, Boucher RC, et al. (July 1998). "An apical PDZ poly peptide anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton". The Journal of Biological Chemistry. 273 (31): 19797–801. doi:10.1074/jbc.273.31.19797. PMID 9677412.
- ^ a b c Enuka Y, Hanukoglu I, Edelheit O, Vaknine H, Hanukoglu A (March 2012). "Epithelial sodium channels (ENaC) are uniformly distributed on motile cilia in the oviduct and the respiratory airways". Histochemistry and Cell Biology. 137 (3): 339–53. doi:10.1007/s00418-011-0904-1. PMID 22207244. S2CID 15178940.
- ^ Sharma Southward, Hanukoglu A, Hanukoglu I (April 2018). "Localization of epithelial sodium channel (ENaC) and CFTR in the germinal epithelium of the testis, Sertoli cells, and spermatozoa". Periodical of Molecular Histology. 49 (2): 195–208. doi:10.1007/s10735-018-9759-2. PMID 29453757. S2CID 3761720.
- ^ Sharma S, Hanukoglu I (Apr 2019). "Mapping the sites of localization of epithelial sodium channel (ENaC) and CFTR in segments of the mammalian epididymis". Journal of Molecular Histology. fifty (2): 141–154. doi:ten.1007/s10735-019-09813-3. PMID 30659401. S2CID 58026884.
- ^ "CF Study Finds New Cells Called Ionocytes Conveying Loftier levels of CFTR Gene". Cystic Fibrosis News Today. three August 2018.
- ^ Montoro DT, Haber AL, Biton M, Vinarsky 5, Lin B, Birket SE, et al. (August 2018). "A revised airway epithelial hierarchy includes CFTR-expressing ionocytes". Nature. 560 (7718): 319–324. Bibcode:2018Natur.560..319M. doi:x.1038/s41586-018-0393-seven. PMC6295155. PMID 30069044.
- ^ Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma Grand, et al. (August 2018). "A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte". Nature. 560 (7718): 377–381. Bibcode:2018Natur.560..377P. doi:10.1038/s41586-018-0394-six. PMC6108322. PMID 30069046.
- ^ a b Hanukoglu I, Boggula VR, Vaknine H, Sharma Due south, Kleyman T, Hanukoglu A (June 2017). "Expression of epithelial sodium channel (ENaC) and CFTR in the human epidermis and epidermal appendages". Histochemistry and Prison cell Biology. 147 (6): 733–748. doi:x.1007/s00418-016-1535-3. PMID 28130590. S2CID 8504408.
- ^ Moskwa P, Lorentzen D, Excoffon KJ, Zabner J, McCray Atomic number 82, Nauseef WM, et al. (January 2007). "A novel host defense arrangement of airways is lacking in cystic fibrosis". American Journal of Respiratory and Critical Care Medicine. 175 (2): 174–83. doi:ten.1164/rccm.200607-1029OC. PMC2720149. PMID 17082494.
- ^ Xu Y, Szép S, Lu Z (Dec 2009). "The antioxidant role of thiocyanate in the pathogenesis of cystic fibrosis and other inflammation-related diseases". Proceedings of the National Academy of Sciences of the Us. 106 (48): 20515–9. Bibcode:2009PNAS..10620515X. doi:ten.1073/pnas.0911412106. PMC2777967. PMID 19918082.
- ^ Yonei Y, Tanaka M, Ozawa Y, Miyazaki Thou, Tsukada North, Inada S, et al. (April 1992). "Primary hepatocellular carcinoma with severe hypoglycemia: involvement of insulin-like growth factors". Liver. 12 (2): ninety–3. doi:x.1111/j.1600-0676.1992.tb00563.x. PMID 1320177.
- ^
- ^ Cheng J, Moyer BD, Milewski M, Loffing J, Ikeda M, Mickle JE, et al. (February 2002). "A Golgi-associated PDZ domain protein modulates cystic fibrosis transmembrane regulator plasma membrane expression". The Journal of Biological Chemistry. 277 (v): 3520–9. doi:x.1074/jbc.M110177200. PMID 11707463.
- ^ a b Gentzsch M, Cui L, Mengos A, Chang XB, Chen JH, Riordan JR (February 2003). "The PDZ-binding chloride aqueduct ClC-3B localizes to the Golgi and associates with cystic fibrosis transmembrane conductance regulator-interacting PDZ proteins". The Journal of Biological Chemical science. 278 (eight): 6440–9. doi:10.1074/jbc.M211050200. PMID 12471024.
- ^ Wang Due south, Yue H, Derin RB, Guggino WB, Li M (September 2000). "Accessory poly peptide facilitated CFTR-CFTR interaction, a molecular machinery to potentiate the chloride channel activity". Cell. 103 (ane): 169–79. doi:10.1016/S0092-8674(00)00096-nine. PMID 11051556. S2CID 16697781.
- ^ Liedtke CM, Yun CH, Kyle Due north, Wang D (June 2002). "Protein kinase C epsilon-dependent regulation of cystic fibrosis transmembrane regulator involves binding to a receptor for activated C kinase (RACK1) and RACK1 binding to Na+/H+ exchange regulatory cistron". The Periodical of Biological Chemical science. 277 (25): 22925–33. doi:10.1074/jbc.M201917200. PMID 11956211.
- ^ a b Park One thousand, Ko SB, Choi JY, Muallem G, Thomas PJ, Pushkin A, et al. (Dec 2002). "The cystic fibrosis transmembrane conductance regulator interacts with and regulates the action of the HCO3- save transporter human Na+-HCO3- cotransport isoform iii". The Journal of Biological Chemistry. 277 (52): 50503–9. doi:10.1074/jbc.M201862200. PMID 12403779.
- ^ a b Cormet-Boyaka Eastward, Di A, Chang SY, Naren AP, Tousson A, Nelson DJ, Kirk KL (September 2002). "CFTR chloride channels are regulated by a SNAP-23/syntaxin 1A complex". Proceedings of the National Academy of Sciences of the United States of America. 99 (19): 12477–82. Bibcode:2002PNAS...9912477C. doi:10.1073/pnas.192203899. PMC129470. PMID 12209004.
- ^ Hegedüs T, Sessler T, Scott R, Thelin West, Bakos East, Váradi A, et al. (March 2003). "C-terminal phosphorylation of MRP2 modulates its interaction with PDZ proteins". Biochemical and Biophysical Enquiry Communications. 302 (3): 454–61. doi:10.1016/S0006-291X(03)00196-seven. PMID 12615054.
- ^ Wang S, Raab RW, Schatz PJ, Guggino WB, Li Grand (May 1998). "Peptide binding consensus of the NHE-RF-PDZ1 domain matches the C-terminal sequence of cystic fibrosis transmembrane conductance regulator (CFTR)". FEBS Letters. 427 (1): 103–8. doi:10.1016/S0014-5793(98)00402-v. PMID 9613608. S2CID 20803242.
- ^ Moyer BD, Duhaime Chiliad, Shaw C, Denton J, Reynolds D, Karlson KH, et al. (September 2000). "The PDZ-interacting domain of cystic fibrosis transmembrane conductance regulator is required for functional expression in the upmost plasma membrane". The Periodical of Biological Chemistry. 275 (35): 27069–74. doi:10.1074/jbc.M004951200. PMID 10852925.
- ^ Hall RA, Ostedgaard LS, Premont RT, Blitzer JT, Rahman N, Welsh MJ, Lefkowitz RJ (July 1998). "A C-terminal motif found in the beta2-adrenergic receptor, P2Y1 receptor and cystic fibrosis transmembrane conductance regulator determines bounden to the Na+/H+ exchanger regulatory factor family unit of PDZ proteins". Proceedings of the National Academy of Sciences of the U.s.a. of America. 95 (15): 8496–501. Bibcode:1998PNAS...95.8496H. doi:10.1073/pnas.95.fifteen.8496. PMC21104. PMID 9671706.
- ^ Lord's day F, Hug MJ, Lewarchik CM, Yun CH, Bradbury NA, Frizzell RA (September 2000). "E3KARP mediates the association of ezrin and protein kinase A with the cystic fibrosis transmembrane conductance regulator in airway cells". The Periodical of Biological Chemistry. 275 (38): 29539–46. doi:x.1074/jbc.M004961200. PMID 10893422.
- ^ Naren AP, Nelson DJ, Xie Due west, Jovov B, Pevsner J, Bennett MK, et al. (November 1997). "Regulation of CFTR chloride channels by syntaxin and Munc18 isoforms". Nature. 390 (6657): 302–five. Bibcode:1997Natur.390..302N. doi:10.1038/36882. PMID 9384384. S2CID 4395005.
- ^ Egan ME (March 2016). "Genetics of Cystic Fibrosis: Clinical Implications". Clinics in Breast Medicine. 37 (1): 9–16. doi:10.1016/j.ccm.2015.11.002. PMID 26857764.
- ^ De Boeck K, Amaral MD (August 2016). "Progress in therapies for cystic fibrosis". The Lancet. Respiratory Medicine. 4 (8): 662–674. doi:10.1016/S2213-2600(sixteen)00023-0. PMID 27053340.
- ^ Thiagarajah JR, Verkman As (September 2012). "CFTR inhibitors for treating diarrheal disease". Clinical Pharmacology and Therapeutics. 92 (three): 287–90. doi:10.1038/clpt.2012.114. PMC3643514. PMID 22850599.
- ^ Jones AM, Helm JM (Oct 2009). "Emerging treatments in cystic fibrosis". Drugs. 69 (14): 1903–10. doi:10.2165/11318500-000000000-00000. PMID 19747007. S2CID 23344660.
- ^ McPhail GL, Clancy JP (April 2013). "Ivacaftor: the first therapy interim on the chief crusade of cystic fibrosis". Drugs of Today. 49 (4): 253–60. doi:10.1358/dot.2013.49.4.1940984. PMID 23616952.
- ^ "Phase three Study of VX-770 Shows Marked Improvement in Lung Function Among People with Cystic Fibrosis with G551D Mutation". Press Release. Cystic Fibrosis Foundation. 2011-02-23.
- ^ Herper M (27 December 2012). "The About Important New Drug Of 2012". Forbes.
- ^ Nocera J (eighteen July 2014). "The $300,000 Drug". The New York Times.
Further reading [edit]
- Kulczycki LL, Kostuch M, Bellanti JA (January 2003). "A clinical perspective of cystic fibrosis and new genetic findings: relationship of CFTR mutations to genotype-phenotype manifestations". American Journal of Medical Genetics. Function A. 116A (3): 262–seven. doi:10.1002/ajmg.a.10886. PMID 12503104. S2CID 9245855.
- Vankeerberghen A, Cuppens H, Cassiman JJ (March 2002). "The cystic fibrosis transmembrane conductance regulator: an intriguing protein with pleiotropic functions". Journal of Cystic Fibrosis. 1 (1): thirteen–29. doi:10.1016/S1569-1993(01)00003-0. PMID 15463806.
- Tsui LC (1992). "Mutations and sequence variations detected in the cystic fibrosis transmembrane conductance regulator (CFTR) gene: a report from the Cystic Fibrosis Genetic Analysis Consortium". Human being Mutation. 1 (3): 197–203. doi:x.1002/humu.1380010304. PMID 1284534. S2CID 35904538.
- McIntosh I, Cutting GR (July 1992). "Cystic fibrosis transmembrane conductance regulator and the etiology and pathogenesis of cystic fibrosis". FASEB Journal. 6 (10): 2775–82. doi:10.1096/fasebj.6.x.1378801. PMID 1378801. S2CID 24932803.
- Drumm ML, Collins FS (1993). "Molecular biology of cystic fibrosis". Molecular Genetic Medicine. 3: 33–68. doi:10.1016/b978-0-12-462003-2.50006-7. ISBN9780124620032. PMID 7693108.
- Kerem B, Kerem E (1996). "The molecular footing for disease variability in cystic fibrosis". European Journal of Human Genetics. 4 (2): 65–73. doi:ten.1159/000472174. PMID 8744024. S2CID 41476164.
- Devidas S, Guggino WB (Oct 1997). "CFTR: domains, structure, and function". Journal of Bioenergetics and Biomembranes. 29 (5): 443–51. doi:10.1023/A:1022430906284. PMID 9511929. S2CID 6000695.
- Nagel M (December 1999). "Differential function of the two nucleotide binding domains on cystic fibrosis transmembrane conductance regulator". Biochimica et Biophysica Acta (BBA) - Biomembranes. 1461 (2): 263–74. doi:10.1016/S0005-2736(99)00162-five. PMID 10581360.
- Boyle MP (2000). "Unique presentations and chronic complications in adult cystic fibrosis: do they teach united states annihilation about CFTR?". Respiratory Research. 1 (3): 133–v. doi:ten.1186/rr23. PMC59552. PMID 11667976.
- Greger R, Schreiber R, Mall K, Wissner A, Hopf A, Briel Grand, et al. (2001). "Cystic fibrosis and CFTR". Pflügers Archiv. 443 Suppl i: S3-7. doi:x.1007/s004240100635. PMID 11845294. S2CID 8057614.
- Bradbury NA (2001). "campsite signaling cascades and CFTR: is there more to learn?". Pflügers Archiv. 443 Suppl 1: S85-91. doi:10.1007/s004240100651. PMID 11845310. S2CID 19373036.
- Dahan D, Evagelidis A, Hanrahan JW, Hinkson DA, Jia Y, Luo J, Zhu T (2001). "Regulation of the CFTR channel by phosphorylation". Pflügers Archiv. 443 Suppl one: S92-six. doi:10.1007/s004240100652. PMID 11845311. S2CID 8144727.
- Cohn JA, Noone PG, Jowell PS (September 2002). "Idiopathic pancreatitis related to CFTR: circuitous inheritance and identification of a modifier factor". Journal of Investigative Medicine. l (five): 247S–255S. doi:10.1136/jim-50-suppl5-01. PMID 12227654. S2CID 34017638.
- Schwartz Yard (February 2003). "[Cystic fibrosis transmembrane conductance regulator (CFTR) gene: mutations and clinical phenotypes]". Ugeskrift for Laeger. 165 (9): 912–half dozen. PMID 12661515.
- Wong LJ, Alper OM, Wang BT, Lee MH, Lo SY (July 2003). "Two novel null mutations in a Taiwanese cystic fibrosis patient and a survey of East Asian CFTR mutations". American Journal of Medical Genetics. Part A. 120A (2): 296–eight. doi:x.1002/ajmg.a.20039. PMID 12833420. S2CID 41060230.
- Cuppens H, Cassiman JJ (October 2004). "CFTR mutations and polymorphisms in male infertility". International Journal of Andrology. 27 (five): 251–6. doi:10.1111/j.1365-2605.2004.00485.10. PMID 15379964.
- Cohn JA, Mitchell RM, Jowell PS (March 2005). "The impact of cystic fibrosis and PSTI/SPINK1 gene mutations on susceptibility to chronic pancreatitis". Clinics in Laboratory Medicine. 25 (ane): 79–100. doi:10.1016/j.cll.2004.12.007. PMID 15749233.
- Southern KW, Peckham D (2004). "Establishing a diagnosis of cystic fibrosis". Chronic Respiratory Illness. one (4): 205–10. doi:10.1191/1479972304cd044rs. PMID 16281647.
- Kandula L, Whitcomb DC, Lowe ME (June 2006). "Genetic problems in pediatric pancreatitis". Electric current Gastroenterology Reports. eight (3): 248–53. doi:10.1007/s11894-006-0083-8. PMID 16764792. S2CID 23606613.
- Marcet B, Boeynaems JM (December 2006). "Relationships between cystic fibrosis transmembrane conductance regulator, extracellular nucleotides and cystic fibrosis". Pharmacology & Therapeutics. 112 (3): 719–32. doi:10.1016/j.pharmthera.2006.05.010. PMID 16828872.
- Wilschanski 1000, Durie PR (August 2007). "Patterns of GI disease in adulthood associated with mutations in the CFTR gene". Gut. 56 (eight): 1153–63. doi:10.1136/gut.2004.062786. PMC1955522. PMID 17446304.
External links [edit]
- GeneReviews/NCBI/NIH/UW entry on CFTR-Related Disorders - Cystic Fibrosis (CF, Mucoviscidosis) and Congenital Absence of the Vas Deferens (CAVD)
- The Cystic Fibrosis Transmembrane Conductance Regulator Protein
- The Human Gene Mutation Database - CFTR Records
- Cystic Fibrosis Mutation Database
- Oak Ridge National Laboratory CFTR Information
- CFTR at OMIM (National Center for Biotechnology Information)
- Overview of all the structural information available in the PDB for UniProt: P13569 (Human Cystic fibrosis transmembrane conductance regulator) at the PDBe-KB.
- Overview of all the structural information bachelor in the PDB for UniProt: P26361 (Mouse Cystic fibrosis transmembrane conductance regulator) at the PDBe-KB.
Source: https://en.wikipedia.org/wiki/Cystic_fibrosis_transmembrane_conductance_regulator




0 Response to "what happens to the chloride gradient in CF"
Post a Comment